Using Pandas as a Unified IO Tool

When I wrote my dissertation, I used Pandoc to convert my draft in markdown format to the final version in PDF format. Pandoc is an extremely powerful and easy-to-use tool for file conversion, as shown in the following diagram.
When we load data into Python, we have a similar demand as well: using a unified and powerful tool to read / write data. From last year, I have dived into Pandas, and considered it as such a perfect candidate.
According to the newest Pandas doc, Pandas supports reading and supporting these commonly-used file format: CSV, JSON, HTML, Local clipboard, MS Excel, HDF5 Format, Feather Format, Msgpack, Stata, SAS, Python Pickle Format, SQL, and Google Big Query. If we visualize these data formats, we can have a clearer idea:
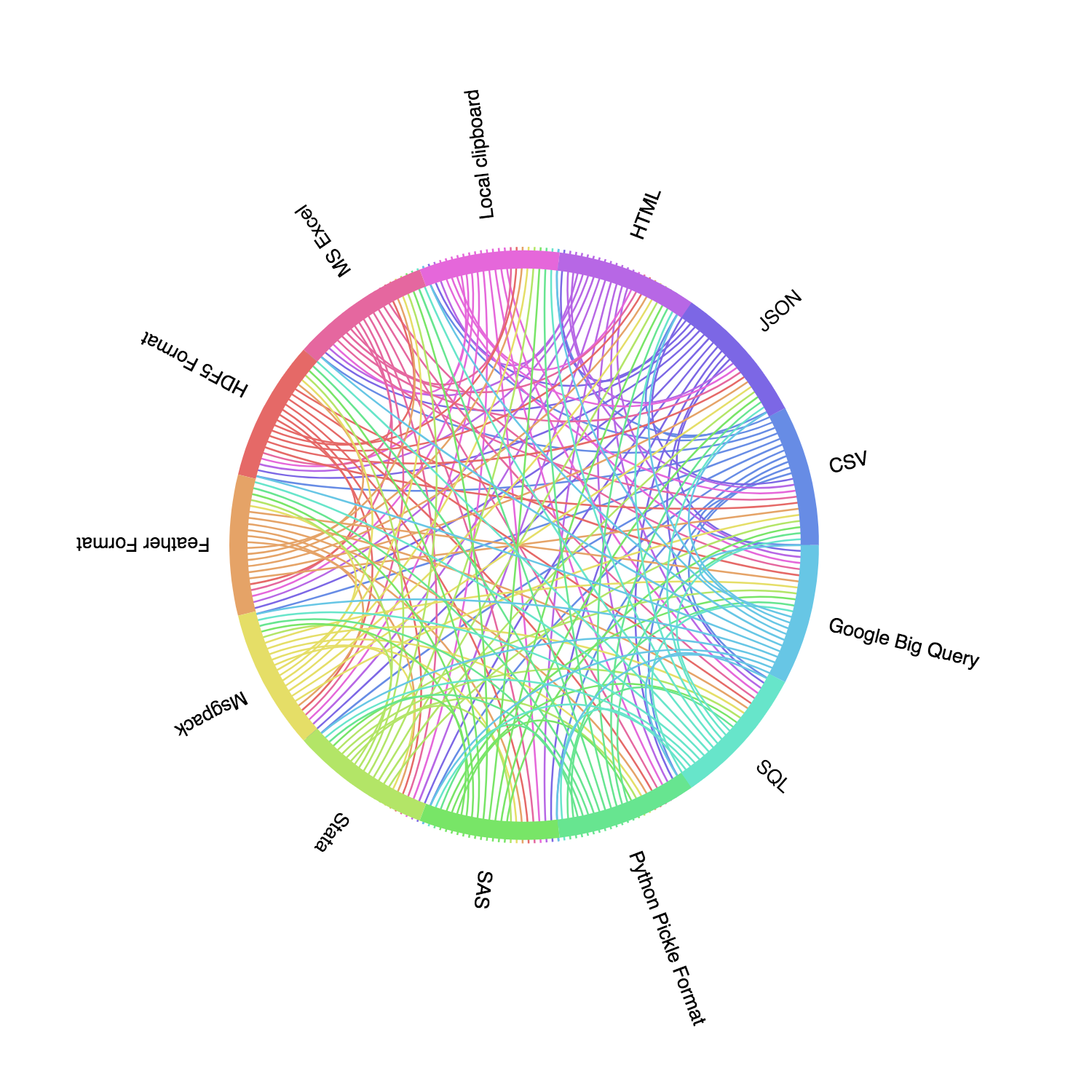
A comprehensive introduction of Pandas IO tools can be found here. However, in this post, we will briefly introduce using Pandas to read / write some common file format.
CSV
CSV (comma-separated-value) format is one of the most common formats in data processing. It is easy for both human and machine to read.
data = pd.read_csv(in_file, quote=0, sep=',', engine='c')quote is to tell which quotation convention the data uses.
If the sep set as None and engine as ‘python’, this function will automatically sniff the delimiter.
c engine is much faster (at least 50%) than python engine, but python engine supports more features
data.to_csv(out_file, header=True, index=False)
If we want to keep header and index, we can set `header` and `index` as `True`, and vice versa.TSV
TSV (tab-separated-value) format is also very common, and Pandas can process it in a similar way as CSV.
data = pd.read_table(in_file, quote=0, sep='\t', engine='c')JSON
JSON has gain more popularity recently. It has more controls on data, but it is not very human-friendly. Because it has a number of orients, it is quite easy to get confused. Therefore, when we use Pandas to read a JSON file, we have to specify the orient. It could be split, records, index, columns or values. Moreover, it the file is line-based, we can set lines as True.
data = pd.read_json(in_file, orient='records', lines=False)data.to_json(out_file, orient='records', lines=False)HDF5
HDF5 is a unique file format. We can include multiple other-format files into a single HDF5 file, and used a key to index them. Therefore, we can save space and reading speed of multiple files.
def hdf2df(in_hdf, hdf_keys):
"""
Read a hdf5 file and return all dfs
:param in_hdf: a hdf5 file
:param hdf_keys:
:return a dict of df
"""
return {i: pd.read_hdf(in_hdf, i) for i in hdf_keys}def df2hdf(out_hdf, dfs, hdf_keys, mode='a'):
"""
Store single or multiple dfs to one hdf5 file
:param dfs: single of multiple dfs
:param out_hdf: the output file
:param hdf_keys: [key for hdf]
"""
for j, k in zip(dfs, hdf_keys):
j.to_hdf(out_hdf, k, table=True, mode=mode)MySQL
MySQL is one of the most popular database, and pandas can easily read the data from it with the help of another Python library sqlalchemy.
First, we use sqlalchemy to make a MySQL connection.
from sqlalchemy import create_engine
def connect_db(host):
return create_engine(host)Then, we give a SQL query to pandas, and query from the created connection. Just that simple, we can easiily get the queried result to a pandas Dataframe.
def mysql_df(sql, con):
df = pd.read_sql_query(sql=sql, con=con)
return dfAdvantages
Using Pandas as a unified IO tool has two main advantages:
1. Pandas IO tools provide a significant performance increase when reading or writing data.
2. Pandas has very detailed document, so the learning curse is reduced.
Therefore, I strongly encourage you to use Pandas a unified IO tool for data reading / writing.



Comments ()